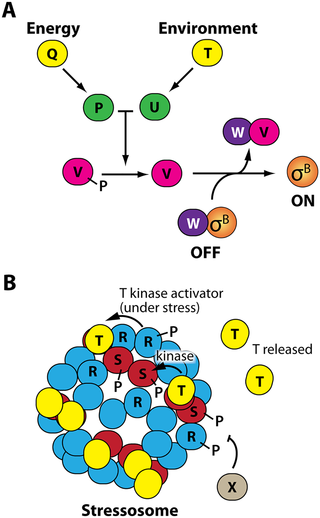
Because of their small size, bacteria experience the presence of stressors in their local environments virtually instantaneously. This means that bacteria must have systems in place that sense stress and are poised to respond very rapidly but also with an appropriate magnitude and duration. As in eukaryotic systems such as the NF-κB stress-response system, a rapid response is ensured by making a stress-response protein ahead of time and then holding it in an inactive state until a stress signal is received. In the model bacterium B. subtilis, the stress-response protein is a sigma factor known as σB. Sigma factors are proteins that tell RNA polymerase where to bind to DNA and transcribe (express) genes. When σB is active, it directs the transcription of a group of some 150 stress-response genes that confer a so-called "general stress response" that helps cells survive the sudden onset of many different types of stress. However, most of the specific functions of the general-stress-response proteins remain unknown.
When cells are not under stress, σB is kept in an inactive state by an anti-sigma factor called RsbV (or just "V" for simplicity). V binds to σB and prevents it from bringing RNA polymerase to the general stress-response genes. When cells sense stress, a signaling cascade is initiated (figure) that leads to the dephosphorylation (i.e., removal of a phosphate group) of V, which makes it let go of σB and instead bind an anti-anti-sigma factor, RsbW ("W"). Now σB is free to activate the general stress response.
The upstream signaling cascades are illustrated in the figure. Energy or nutritional stress, sensed as the depletion of ATP (the cellular energy currency), is sensed through an unknown mechanism by RsbQ and RsbP—P is a phosphatase that can dephosphorylate V. On the other hand, environmental stress—ethanol, heat, acid, salt, antibiotics, etc.—is sensed by large 80-protein complexes called stressosomes. Stressosomes are composed of RsbR and RsbS proteins, and they also bind to a protein called RsbT. Environmental stressors cause stressosomes to release T in a process that involves T phosphorylating R and S, and the free T then activates RsbU—U is another phosphatase that can dephosphorylate V.
Further complicating things is that the stressosome contains a mixture of four "flavors" (paralogs) of R proteins that are similar but distinct. Why does B. subtilis make these similar but different proteins? Our work thus far has taken advantage of microfluidic technology to uncover differences among these R proteins in their stress-response profiles when responding to ethanol, a classic stressor. We now seek to discover the molecular bases of these differences and to understand how different stress-response profiles can confer survival advantages to cells.
Here are some of the fundamental questions we are asking:
What is actually sensed by the R proteins in the stressosome?
Do the different flavors of R proteins respond differently to different stresses?
What parts of the R proteins govern their distinct response profiles?
Which amino-acid residues play key roles in stress sensing by R proteins?
How do different combinations of R proteins affect overall response profiles?
How do different response profiles confer survival advantages or disadvantages in the presence of different stresses?
As you can see, we have many promising avenues of research. Understanding how different stress-response profiles are generated and how they influence cell survival will give greater insights into the strategies that cells have evolved to maximize their fitness while being prepared for ever-changing conditions.
We are making gains in these areas--please see our Publications page to see our most recent work!
Interested in working on these projects? Contact us—we are always looking for motivated students who love to learn.